1. Model Introduction
K-Means is one of the most commonly used clustering algorithms in unsupervised machine learning. It divides a given data set into multiple different groups or clusters. The core point of each group is the mean value of the group members. Points (usually called “centroids”) can be represented as multidimensional vectors.
The goal of K-Means algorithm is to find groups in the data, minimizing the sum of squared distances from group members to the actual centroid. With the continuous increase of the analyzed data set, it is necessary to calculate the distance between all data points and the centroid each time. To tackle the increased calculation amount and time, the Mini Batch K-Means method is adopted. Mini Batch K-Means is a variant of K-Means. The difference is that Mini Batch K-Means only needs to sample and iterate from all data sets each time, shortening the calculation time to a large degree. It is also widely used to reduce the training time of the algorithm in the Back-propagation.
2. R&D Bases
(1) Béjar,Javoer. K-means vs Mini Batch K-Means: A comparison. Tech. rep. Universitat Politècnica de Catalunya (2013).
(2) HAN Pu1,WANG Dongbo, LIU Yanyun, SU Xinning. The Influence of Speeches on Chinese and English Document Clustering. Journal of Chinese Information Processing,2013,27(2):65-73.
3. Algorithm Description
Randomly select some samples from the data set to form a mini batch; assign them to the nearest group, and continuously update and adjust the centroid. When the convergence conditions (such as the number of iterations, accuracy) are met, the calculation can be terminated.
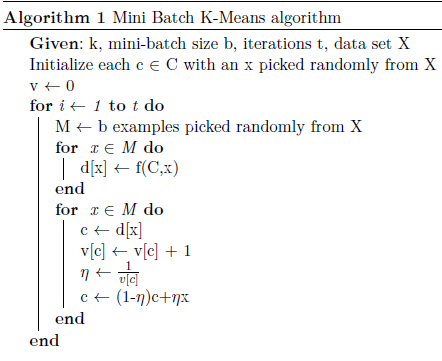
Mini Batch K-Means saves a lot of calculation time compared to K-Means. Although a small amount of clustering quality will be sacrificed, the difference in accuracy is negligible when the amount of data is bigger.
4. Index Evaluation
(1) The sum of squared errors within the cluster: the inertial pointer in the K-Means method represents the sum of the mean square distance between each sample and the centroid of the cluster. Generally, the smaller the inertia, the better the model. But as K value increases, the rate of inertia decline becomes very slow, so the K value of the “elbow” can be selected as the most suitable K value choice.
(2) Contour coefficient: the calculation method is that when the average distance between the sample and other samples in the cluster is a, and the average distance between the sample and the external cluster sample is b, the contour coefficient is (b-a)/max(a,b). Generally, the larger the contour coefficient index, the better the clustering effect.
